
Create an interactive loon plot widget
Source:R/l_plot.R, R/l_plot_decomposed_ts.R, R/l_plot_density.R, and 2 more
l_plot.Rdl_plot is a generic function for creating an interactive
visualization environments for R objects.
Usage
l_plot(x, y, ...)
# Default S3 method
l_plot(
x,
y = NULL,
by = NULL,
on,
layout = c("grid", "wrap", "separate"),
connectedScales = c("cross", "row", "column", "both", "x", "y", "none"),
color = l_getOption("color"),
glyph = l_getOption("glyph"),
size = l_getOption("size"),
active = TRUE,
selected = FALSE,
xlabel,
ylabel,
title,
showLabels = TRUE,
showScales = FALSE,
showGuides = TRUE,
guidelines = l_getOption("guidelines"),
guidesBackground = l_getOption("guidesBackground"),
foreground = l_getOption("foreground"),
background = l_getOption("background"),
parent = NULL,
...
)
# S3 method for class 'decomposed.ts'
l_plot(
x,
y = NULL,
xlabel = NULL,
ylabel = NULL,
title = NULL,
tk_title = NULL,
color = l_getOption("color"),
size = l_getOption("size"),
linecolor = l_getOption("color"),
linewidth = l_getOption("linewidth"),
linkingGroup,
showScales = TRUE,
showGuides = TRUE,
showLabels = TRUE,
...
)
# S3 method for class 'density'
l_plot(
x,
y = NULL,
xlabel = NULL,
ylabel = NULL,
title = NULL,
linewidth = l_getOption("linewidth"),
linecolor = l_getOption("color"),
...
)
# S3 method for class 'map'
l_plot(x, y = NULL, ...)
# S3 method for class 'stl'
l_plot(
x,
y = NULL,
xlabel = NULL,
ylabel = NULL,
title = NULL,
tk_title = NULL,
color = l_getOption("color"),
size = l_getOption("size"),
linecolor = l_getOption("color"),
linewidth = l_getOption("linewidth"),
linkingGroup,
showScales = TRUE,
showGuides = TRUE,
showLabels = TRUE,
...
)Arguments
- x
the coordinates of points in the
l_plot. Alternatively, a single plotting structure (see the functionxy.coordsfor details),formula, or any R object (e.g.density,stl, etc) is accommodated.- y
the y coordinates of points in the
l_plot, optional if x is an appropriate structure.- ...
named arguments to modify plot states. See
l_info_statesof any instantiatedl_plotfor examples of names and values.- by
loon plot can be separated by some variables into multiple panels. This argument can take a
formula,ndimensional state names (seel_nDimStateNames) ann-dimensionalvectoranddata.frameor alistof same lengthsnas input.- on
if the
xorbyis a formula, an optional data frame containing the variables in thexorby. If the variables are not found in data, they are taken from environment, typically the environment from which the function is called.- layout
layout facets as
'grid','wrap'or'separate'- connectedScales
Determines how the scales of the facets are to be connected depending on which
layoutis used. For each value oflayout, the scales are connected as follows:layout = "wrap":Across all facets, whenconnectedScalesis"x", then only the "x" scales are connected"y", then only the "y" scales are connected"both", both "x" and "y" scales are connected"none", neither "x" nor "y" scales are connected. For any other value, only the "y" scale is connected.
layout = "grid":Across all facets, whenconnectedScalesis"cross", then only the scales in the same row and the same column are connected"row", then both "x" and "y" scales of facets in the same row are connected"column", then both "x" and "y" scales of facets in the same column are connected"x", then all of the "x" scales are connected (regardless of column)"y", then all of the "y" scales are connected (regardless of row)"both", both "x" and "y" scales are connected in all facets"none", neither "x" nor "y" scales are connected in any facets.
- color
colours of points; colours are repeated until matching the number points. Default is found using
l_getOption("color").- glyph
the visual representation of the point. Argument values can be any of
the string names of primitive glyphs:
circles: "circle", "ccircle", "ocircle";squares or boxes: "square", "csquare", "osquare";triangles: "triangle", "ctriangle", "otriangle";diamonds: "diamond", "cdiamond", or "odiamond".
l_primitiveGlyphs().the string names of constructed glyphs:
text as glyphs: seel_glyph_add_text()point ranges: seel_glyph_add_pointrange()polygons: seel_glyph_add_polygon()parallel coordinates: seel_glyph_add_serialaxes()star or radial axes: seel_glyph_add_serialaxes()or any plot created using R: seel_make_glyphs()
- size
size of the symbol (roughly in terms of area). Default is found using
l_getOption("size").- active
a logical determining whether points appear or not (default is
TRUEfor all points). If a logical vector is given of length equal to the number of points, then it identifies which points appear (TRUE) and which do not (FALSE).- selected
a logical determining whether points appear selected at first (default is
FALSEfor all points). If a logical vector is given of length equal to the number of points, then it identifies which points are (TRUE) and which are not (FALSE).- xlabel
Label for the horizontal (x) axis. If missing, one will be inferred from
xif possible.- ylabel
Label for the vertical (y) axis. If missing, one will be inferred from
y(orx) if possible.- title
Title for the plot, default is an empty string.
- showLabels
logical to determine whether axes label (and title) should be presented.
- showScales
logical to determine whether numerical scales should be presented on both axes.
- showGuides
logical to determine whether to present background guidelines to help determine locations.
- guidelines
colour of the guidelines shown when
showGuides = TRUE. Default is found usingl_getOption("guidelines").- guidesBackground
colour of the background to the guidelines shown when
showGuides = TRUE. Default is found usingl_getOption("guidesBackground").- foreground
foreground colour used by all other drawing. Default is found using
l_getOption("foreground").- background
background colour used for the plot. Default is found using
l_getOption("background").- parent
a valid Tk parent widget path. When the parent widget is specified (i.e. not
NULL) then the plot widget needs to be placed using some geometry manager liketkpackortkplacein order to be displayed. See the examples below.- tk_title
provides an alternative window name to Tk's
wm title. IfNULL,stlwill be used.- linecolor
line colour of all time series. Default given by
l_getOption("color").- linewidth
line width of all time series (incl. original and decomposed components. Default given by
l_getOption("linewidth").- linkingGroup
string giving the linkingGroup for all plots. If missing, a default
linkingGroupwill be determined from deparsing the inputx.
Value
The input is a
stlor adecomposed.tsobject, a structure of class"l_ts"containing four loon plots each representing a part of the decomposition by name: "original", "trend", "seasonal", and "remainder"The input is a vector, formula, data.frame, ...
by = NULL: aloonwidget will be returnedbyis notNULL: anl_facetobject (a list) will be returned and each element is aloonwidget displaying a subset of interest.
Details
Like plot in R, l_plot is
the generic plotting function for objects in loon.
The default method l_plot.default produces the interactive
scatterplot in loon.
This is the workhorse of `loon` and is often a key part of many
other displays (e.g. l_pairs and l_navgraph).
For example, the methods include l_plot.default (the basic interactive scatterplot),
l_plot.density (layers output of density in an empty scatterplot),
l_plot.map (layers a map in an empty scatterplot), and
l_plot.stl (a compound display of the output of stl).
A complete list is had from methods(l_plot).
vignette(topic = "introduction", package = "loon")
and to explore loon's website accessible via l_help(). The general direct manipulation and interaction gestures are outlined in the following figures.
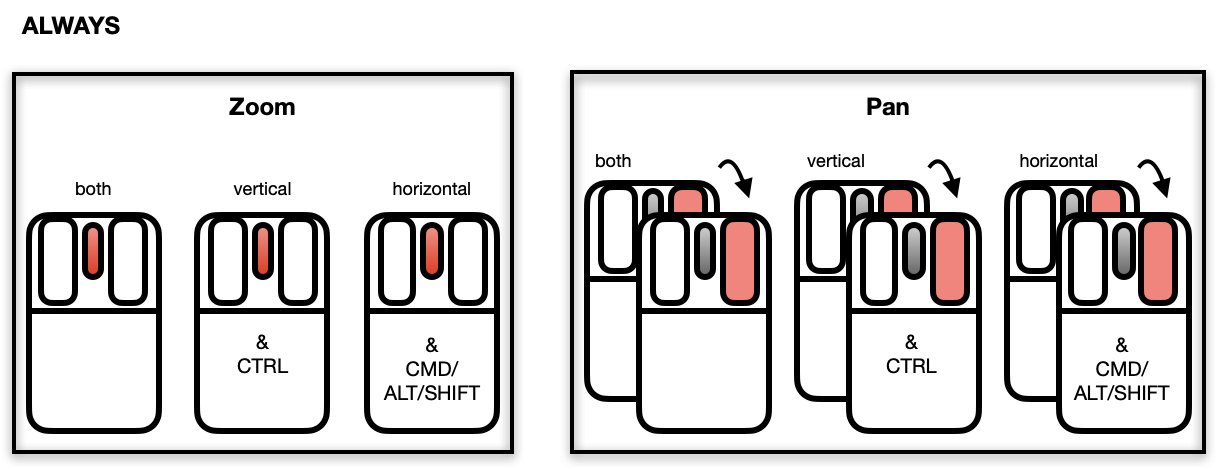
Zooming and Panning
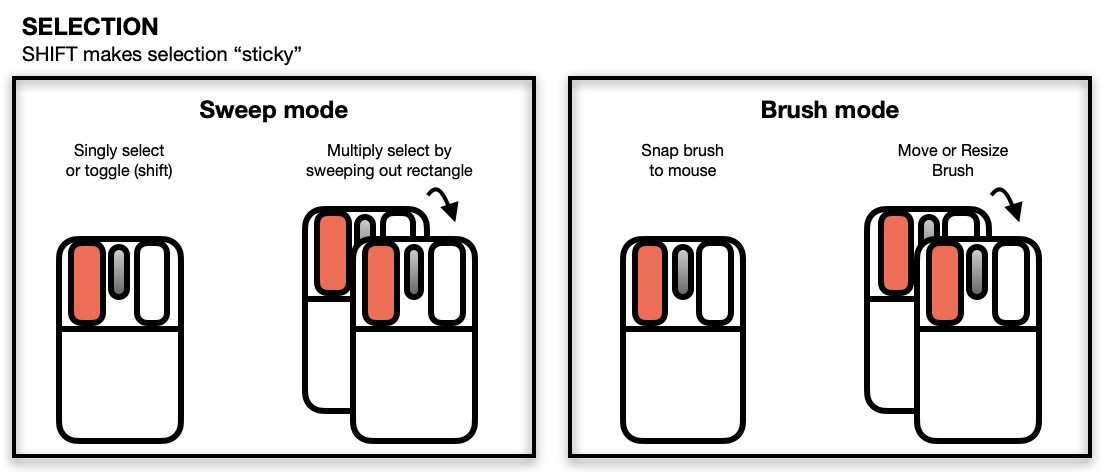
Selecting Points/Objects
Moving Points on the Scatterplot Display
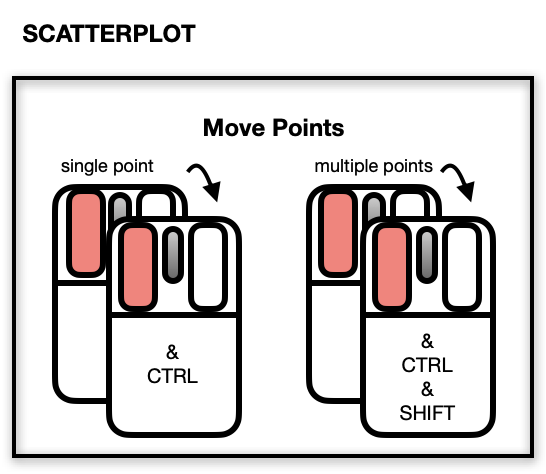
The scatterplot displays a number of direct interactions with the
mouse and keyboard, these include: zooming towards the mouse cursor using
the mouse wheel, panning by right-click dragging and various selection
methods using the left mouse button such as sweeping, brushing and
individual point selection. See the documentation for l_plot
for more details about the interaction gestures.
Some arguments to modify layouts can be passed through,
e.g. "separate", "ncol", "nrow", etc. Check l_facet
to see how these arguments work.
See also
Turn interactive loon plot static loonGrob, grid.loon, plot.loon.
Density layer l_layer.density
Map layer l_layer, l_layer.map,
map
Other loon interactive states:
l_hist(),
l_info_states(),
l_serialaxes(),
l_state_names(),
names.loon()
Examples
if(interactive()) {
########################## l_plot.default ##########################
# default use as scatterplot
p1 <- with(iris, l_plot(Sepal.Length, Sepal.Width, color=Species,
title = "First plot"))
# The names of the info states that can be
# accessed or set. They can also be given values as
# arguments to l_plot.default()
names(p1)
p1["size"] <- 10
p2 <- with(iris, l_plot(Petal.Length ~ Petal.Width,
linkingGroup="iris_data",
title = "Second plot",
showGuides = FALSE))
p2["showScales"] <- TRUE
# link first plot with the second plot requires
# l_configure to coordinate the synchroniztion
l_configure(p1, linkingGroup = "iris_data", sync = "push")
p1['selected'] <- iris$Species == "versicolor"
p2["glyph"][p1['selected']] <- "cdiamond"
gridExtra::grid.arrange(loonGrob(p1), loonGrob(p2), nrow = 1)
# Layout facets
### facet wrap
p3 <- with(mtcars, l_plot(wt, mpg, by = cyl, layout = "wrap"))
# it is equivalent to
# p3 <- l_plot(mpg~wt, by = ~cyl, layout = "wrap", on = mtcars)
### facet grid
p4 <- l_plot(x = 1:6, y = 1:6,
by = size ~ color,
size = c(rep(50, 2), rep(25, 2), rep(50, 2)),
color = c(rep("red", 3), rep("green", 3)))
# Use with other tk widgets
tt <- tktoplevel()
tktitle(tt) <- "Loon plots with custom layout"
p1 <- l_plot(parent=tt, x=c(1,2,3), y=c(3,2,1))
p2 <- l_plot(parent=tt, x=c(4,3,1), y=c(6,8,4))
tkgrid(p1, row=0, column=0, sticky="nesw")
tkgrid(p2, row=0, column=1, sticky="nesw")
tkgrid.columnconfigure(tt, 0, weight=1)
tkgrid.columnconfigure(tt, 1, weight=1)
tkgrid.rowconfigure(tt, 0, weight=1)
########################## l_plot.decomposed.ts ##########################
decompose <- decompose(co2)
p <- l_plot(decompose, title = "Atmospheric carbon dioxide over Mauna Loa")
# names of plots in the display
names(p)
# names of states associated with the seasonality plot
names(p$seasonal)
# which can be set
p$seasonal['color'] <- "steelblue"
########################## l_plot.stl ##########################
co2_stl <- stl(co2, "per")
p <- l_plot(co2_stl, title = "Atmospheric carbon dioxide over Mauna Loa")
# names of plots in the display
names(p)
# names of states associated with the seasonality plot
names(p$seasonal)
# which can be set
p$seasonal['color'] <- "steelblue"
########################## l_plot.density ##########################
# plot a density estimate
set.seed(314159)
ds <- density(rnorm(1000))
p <- l_plot(ds, title = "density estimate",
xlabel = "x", ylabel = "density",
showScales = TRUE)
########################## l_plot.map ##########################
if (requireNamespace("maps", quietly = TRUE)) {
p <- l_plot(maps::map('world', fill=TRUE, plot=FALSE))
}
}